| MadSci Network: Biochemistry |
Dear Kanishk,
Properly, dynein doesn’t really ‘control’ the ciliary motion in the sense of changing direction or what not, but rather it is the motor that causes the motion. It’s function can be regulated, as any motor can be, by turning it on or off (with ATP), but it cannot, properly, be said to exhibit control over the ciliary motion. Having said that, below is a brief description of the generally mechanism behind ciliary motion and dynein’s role in it.
Cilia (the hairs on mammalian cells that are responsible for the motion of the cells – similar to bacterial flagella) contain structures called axonemes. If you think of an axoneme as a bundle of uncooked spaghetti (I will explain shortly) then the motion of the cilia is caused by the spaghetti on one side sliding up and along the ones on the other side. The elongation of one side results in a bending of the entire bundle (Figure 1). Control of the bending results in movement of the cilia. It is the dynein, regulated by ATP, that causes this sliding by ‘walking’ along the spaghetti strands.
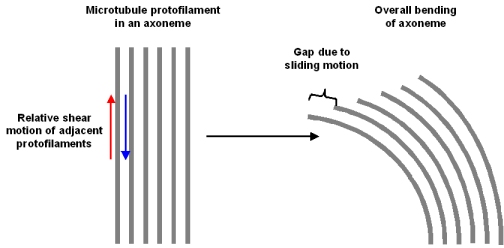
Figure 1: The bending of the axoneme due to sliding of microtubules on one side (from a course web site of, Florida State University Dr. Michael Blaber)
The complexity really arises in regard to the structure and interaction of all the parts of the axoneme (Figure 2). The axoneme (bundle of spaghetti) is composed of many proteins, among these are microtubules (the individual spaghetti strands) and dynein.
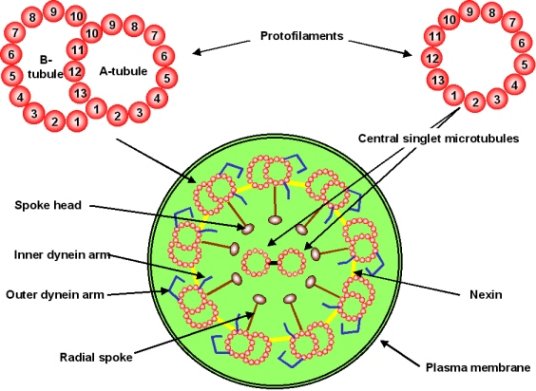
Figure 2: Components of the Axoneme (Dr. Michael Blaber)
Dynein (Figures 3a/b) is a large, multi-component protein that undergoes a change in structure when it binds and hydrolyses ATP. It has several ‘arms’ that bind to microtubules and also bind and hydrolyze ATP.
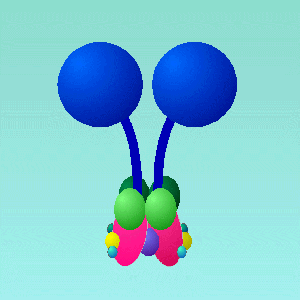
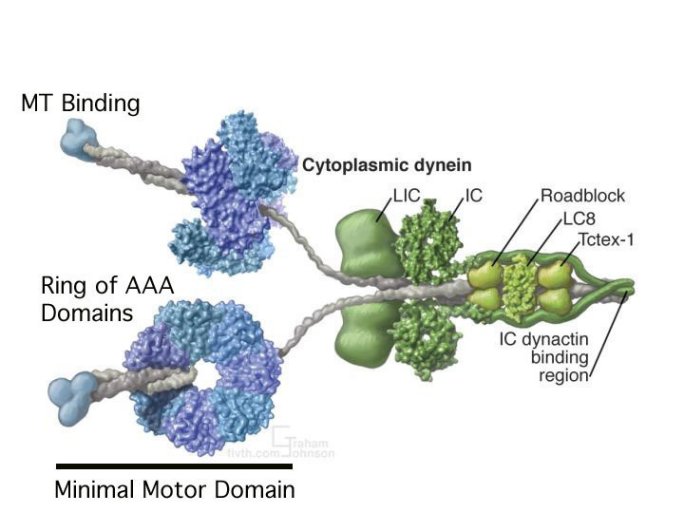
Figure 3: A(top) Schematic representation of Dynein subcomponent arrangement from R.John Lye, University of Virginia. B(lower) Reconstruction of Dynein using actual data of the 3-dimensional structures of some components from The Vale Lab, UCSF
Microtubules (Figure 4) are long tubes composed of many copies of two different proteins (subunits alpha and beta). These subunits are arranged in a helical pattern within the microtubule in such a way that the overall structure has a directionality or ‘polarity’ and is always oriented the same way in the axoneme. In addition, there are two different types of microtubules in the axoneme that differ primarily in the number of subunits in each turn of the helix (they are called A- tubule and B-tubule). The A- and B-tubules are bound like two strands of spaghetti stuck together and there are many of these ‘spaghetti dimers’ arranged around the inner surface of the axoneme.
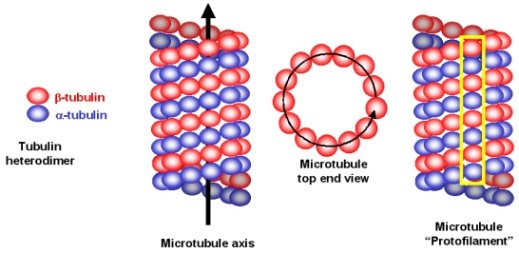
Figure 4: Organisation of microtubules (from Dr. Michael Blaber).
Initially, one arm of dynein binds strongly to A-tubule and its other arm binds weakly to B-tubule. The sliding motion begins when it hydrolyses the ATP that it has been holding (thus creating ADP), it releases the B-tubule and changes conformation to bind the B-tubule in a different place. Dynein then releases the ADP and binds a new ATP, which results in another conformational change to bring the ‘arm’ back to the original position. The result is that the B-tubule has now ‘slid’ up, past the A-tubule. The sliding is regulated so that it occurs first on one side of the axoneme and then the other. Because there are a large number of tubules per axoneme and per cilia that move together in a similar manner, there is a noticeable macroscopic motion (this results in a cilia frequency of 10-40 beats per second).
Interestingly, this dynein-based movement is the same mechanism that allows organelles to move around inside cells (although on cytoskeletal microtubules – sort of like molecular ski lifts).
Unfortunately, the precise molecular details of the dynein conformational changes that induce the motion are not well understood. If you are interested in other natural ‘molecular motors’, the actin/myosin system of muscles has been characterized in more detail.
I hope this has been helpful,
Dr. Edwin Rydberg
Try the links in the MadSci Library for more information on Biochemistry.