| MadSci Network: Molecular Biology |
Hi Uma,
The short answer to your questions is no, not all exons code for proteins.
Exons (as defined by Wikipedia) are the regions of DNA within a gene that are not spliced out from the transcribed RNA and are retained in the final messenger RNA (mRNA) molecule (incidentally, Wikipedia also tells us that the term "exon" was coined by Walter Gilbert in 1978). You will note that there are no comments regarding translation or coding in this definition.
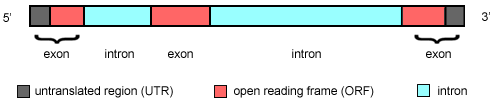
Of course, most exons do code for proteins (or rather, pieces of proteins - it can take more than ten exons to code for some proteins). However, exons have been catagorized into 12 different types, each containing a particular combination of translated and untranslated regions (UTRs, also called nontranslated regions, or NTRs). For example, some of the most common exons with untranslated regions occur at the 5' (beginning) or 3' (end) of the gene and are known 5'- or 3'UTRs. These regions can be important for efficient translation, mRNA stability, or mRNA half-life. Often they will have a very definite secondary structure (a concept usually reserved for proteins) that is crucial for their binding to proteins responsible for initiating translation of the gene. This can be particularly true for viruses. For example, the 5'NTR of Hepatitis C virus contains the highly conserved internal ribosomal entry site (IRES) which is crucial for viral replication and is a target for antiviral therapies.
I hope this helps.
Dr. Edwin H. Rydberg
References:
2)M.Q.Zhang, Statistical features of human exons and their flanking regions. (1998) Human Molecular Genetics 7(5): 919-932.
3) Partho Sarothi Ray and Saumitra Das, Inhibition of hepatitis C virus IRES-mediated translation by small RNAs analogous to stem-loop structures of the 5'-untranslated region. (2004) Nucleic Acids Res. 32(5): 1678ĘC1687.
Try the links in the MadSci Library for more information on Molecular Biology.