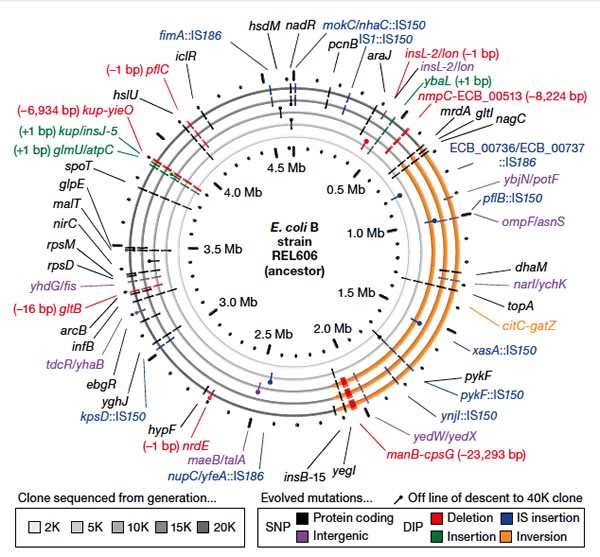
They also followed this (and other strains) for another 20,000 generations, at which
point there were 627 SNPs and 26 insertions, deletions, and inversions. Moreover they found two
surprising results, first they found mutations that increased the mutation rate by about 150 fold,
and then later independent mutations that reduced the high mutation rate by about 50%.
This illustrates the fact that the rate at which mutations occur is itself a selectable trait
(part A - below).
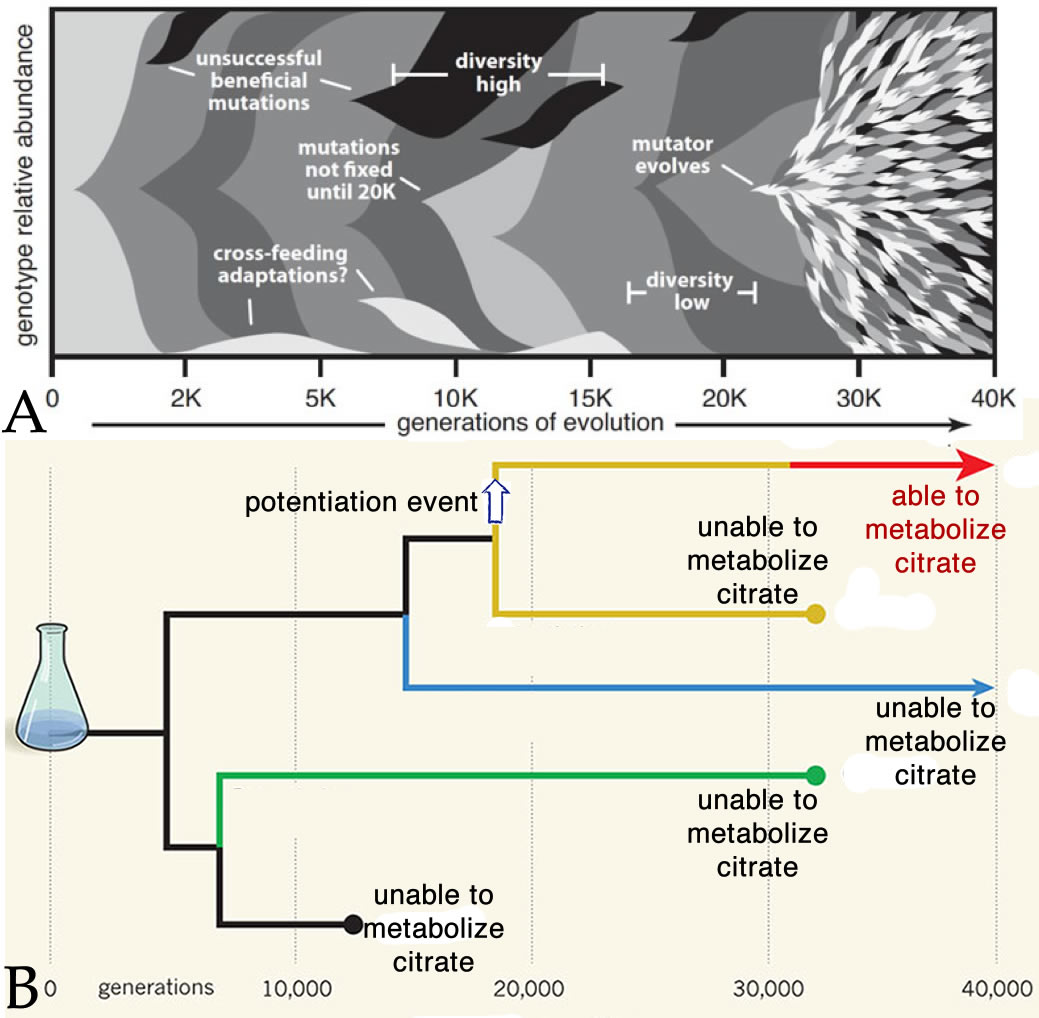
As first noted by Muller in the 1930's (see Henson et al.,
2012), the effects of a mutation on a gene can range from abolishing or reducing its activity
to increasing its activity, to producing a new activity.
The impact of mutations on reproductive success, which is what matters evolutionarily, can
range from the highly negative (such as the inability to make proteins) to the highly positive (such as
the ability to survive in the presence of an antibiotic.) In practice, most mutations are either neutral
in their effects or weakly negative.
Where selection effects are weak, whether a particular mutant allele increases or decreases
in frequency within the population will also be influenced by genetic drift, that is, the stochastic
factors that influence reproductive success.
The other version of the genes (alleles) in a genome influence the effects of a particular
mutation. This was clearly illustrated in the observation that the evolution of the ability to
metabolize citrate was made possible by mutations (potentiation events) that originally had no
dramatic selective benefit in themselves but made the evolution of citrate metabolism by E.
coli possible in later generations. (see Blount et al., 2012)(part
B - above).
Similarly, the origin of the mutator phenotype (around generation 26,000) increased the
ability of cells to develop resistance to various antibiotics. |