Date: Mon Oct 30 17:04:34 2000
Posted By: Michael Onken, MadSci Admin
Area of science: Biochemistry
ID: 972344890.Bc
Message:
Spencer,
Before starting, I would like to clarify some of the terms regarding mRNA:
the "transcription start site" is the site on the DNA, at which RNA
polymerase begins to make the RNA message; the "translation start site" or
"start codon" is the AUG triplet on the mRNA, at which the ribosome begins
to translate the RNA message into a peptide; the "ribosomal binding site" is
the sequence upstream of the start codon, to which the ribosome initially
binds prior to translation. In eukaryotes, there is a 5' cap
consisting of a 5'-5' 7methylGuanidine that can act as the ribosomal binding
site, while most prokaryotes have a conserved Shine-Dalgarno sequence that directs ribosomal
binding. Also, both eukaryotes and prokaryotes use Internal Ribosomal Entry
Sequences (IRES: very common in prokaryotes; very rare in eukaryotes)
that can direct binding of ribosomes to the start codons of multiple
cistrons (genes) on a single message.
So, "what happens if the tag is missing?" If there is no ribosomal binding
site, the mRNA is never translated - this is almost impossible in
eukaryotes, since the 5' cap is added to all messages as part of the post-
transcriptional processing carried out in the nucleus before the mRNA's are
allowed to exit to the cytoplasm. On the other hand, the Shine-Dalgarno
sequence was defined by its requirement for ribosomal binding to cistrons,
so a prokaryotic mRNA without ribosomal binding sites would simply not be
recognized by ribosomes. Also, ribosomes can only read RNA in one
direction, 5' to 3', so it would be impossible for a message to be "run
through backwards," unless it was an "antisense" message.
Your second question was the subject of a Nobel Prize in 1968, and is summarized as
"the Genetic Code:"
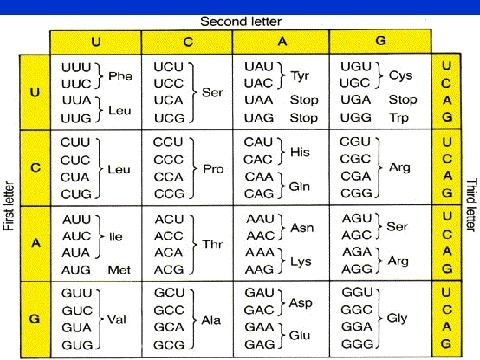
Basically, only two of the twenty common amino acids, methionine and
tryptophan, are coded for by single codons; in most cases there are groups
of two to six codons for each amino acid. One of the more interesting
aspects of the Genetic Code is that most of the groupings of codons involve
variability in the third base of the codon. This has lead to the "Wobble
Hypothesis", that the code may have originally involved fewer amino acids
coded by 16 doublet codons with single base spacers between them. The
theory suggests that as other amino acids became common, the spacers were
used to usurp some of the codons away from the older amino acids (this is
supported to an extent by the biosynthetic pathways of related amino acids).
If you look at the table above, you'll see three stop codons
, UAG, UAA, and UGA. These triplets do not code for amino acids, but
instead tell the ribosome to stop translating the message. Based on early
studies of tRNA's, if there is no tRNA anticodon that recognizes a specific
codon (i.e. the codon does not code for an amino acid), then that codon can
act as a terminator.
Hope this answers your questions, and keep 'em coming!
Michael Onken, MadSci Moderator
Current Queue |
Current Queue for Biochemistry |
Biochemistry archives
Try the links in the MadSci Library for more information on Biochemistry.
MadSci Home | Information |
Search |
Random Knowledge Generator |
MadSci Archives |
Mad Library | MAD Labs |
MAD FAQs |
Ask a ? |
Join Us! |
Help Support MadSci
MadSci Network,
webadmin@www.madsci.org
© 1995-2000. All rights reserved.
